Brucite block in water
We make some includes
from molecular_builder import create_bulk_crystal, carve_geometry, write, pack_water
from molecular_builder.geometry import BoxGeometry
import numpy as np
Create a (too large) block of brucite_in_water
L = np.array([80, 60, 80])
atoms = create_bulk_crystal("brucite", L)
Since create_bulk_crystal makes a block of only whole unit cells, the lengths are probably not exactly L, therefore we update L
L = atoms.cell.lengths()
We then create a geometry and carve out only part of the brucite block.
geometry = BoxGeometry( center=L/2,
length=[L[0], L[1]/2-3, 4*L[2]/5+2],
periodic_boundary_condition=(True, True, True))
carve_geometry(atoms, geometry, side="out")
Compute volumes and pack the right amount of water around the brucite block.
volume = atoms.cell.volume
water_volume = volume-geometry.volume()
pack_water(atoms=atoms, volume=water_volume, pbc=2.0, tolerance=1.8)
Finally, output the system as a LAMMPS data file.
write(atoms, "brucite_in_water.data", bond_specs=("O", "H", 1.02))
In addition, we create two images of the system, one with a perspective projection and one with an orthogonal projection.
write(atoms, "brucite_in_water_perspective.png", bond_specs=("O", "H", 1.02), camera_dir=[1, 0, 0], viewport_type="perspective", size=(480, 640))
write(atoms, "brucite_in_water_orthogonal.png", bond_specs=("O", "H", 1.02), camera_dir=[1, 0, 0], viewport_type="orthogonal", size=(480, 640))
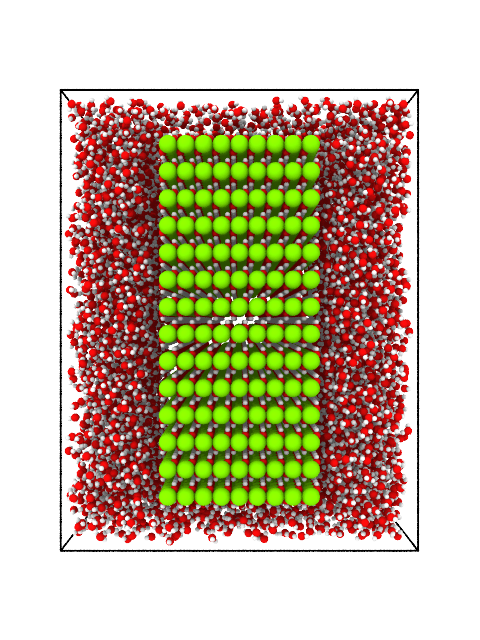
Brucite immersed in water. Perspective projection.
The same image in orthogonal
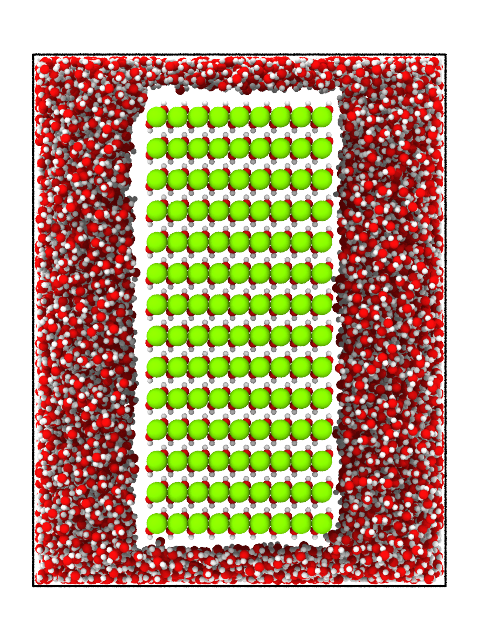
Brucite immersed in water. Orthogonal projection.
The complete script:
from molecular_builder import create_bulk_crystal, carve_geometry, write, pack_water
from molecular_builder.geometry import BoxGeometry
import numpy as np
L = np.array([80, 60, 80])
atoms = create_bulk_crystal("brucite", L)
L = atoms.cell.lengths()
geometry = BoxGeometry( center=L/2,
length=[L[0], L[1]/2-3, 4*L[2]/5+2],
periodic_boundary_condition=(True, True, True))
carve_geometry(atoms, geometry, side="out")
volume = atoms.cell.volume
water_volume = volume-geometry.volume()
pack_water(atoms=atoms, volume=water_volume, pbc=2.0, tolerance=1.8)
write(atoms, "brucite_in_water.data", bond_specs=("O", "H", 1.02))
write(atoms, "brucite_in_water_perspective.png", bond_specs=("O", "H", 1.02), camera_dir=[1, 0, 0], viewport_type="perspective", size=(480, 640))
write(atoms, "brucite_in_water_orthogonal.png", bond_specs=("O", "H", 1.02), camera_dir=[1, 0, 0], viewport_type="orthogonal", size=(480, 640))